An integrated biological and mathematical modeling-based elucidation of asymmetric partitioning of chromosome structural proteins
Project Gist
Understanding how chromosomes self-partition into two functionally-distinct domains after a symmetry-breaking crossover recombination event.
Keywords
Chromosome, Meiosis, Symmetry-breaking, Feedback
Background and Purpose
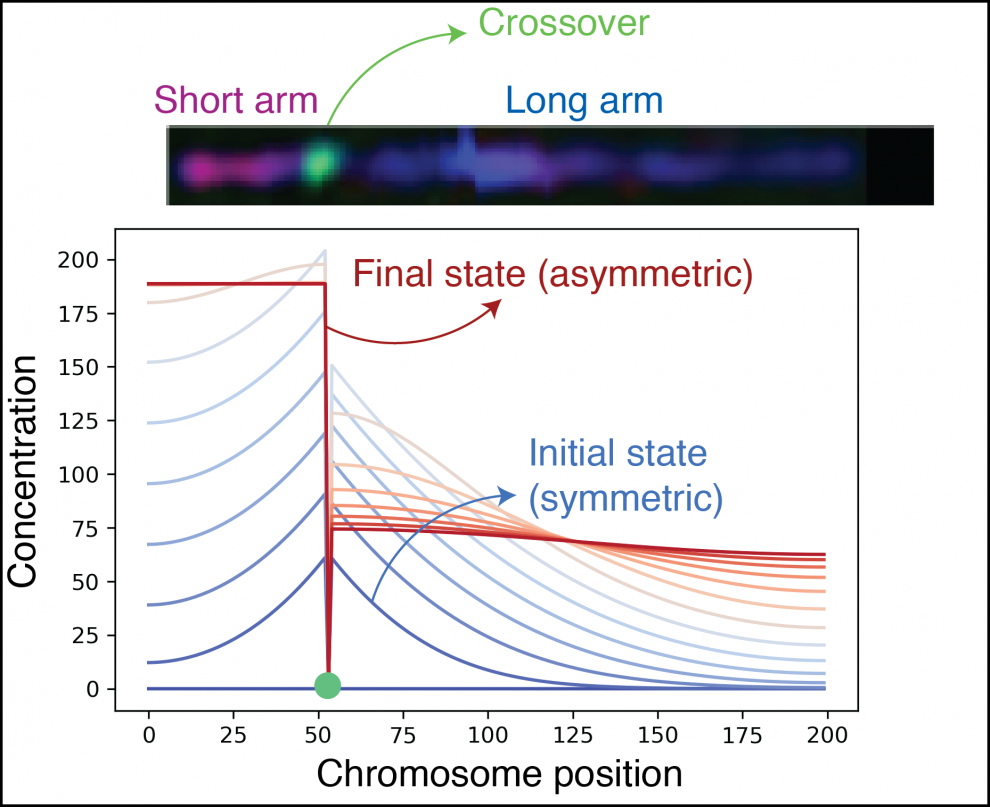
To create haploid gametes, chromosomes need to separate from each other over the course of two divisions in meiosis. The first division takes a group of four linked DNA molecules and divides it into two pairs of two molecules; these pairs stay connected to each other until the second division. In the nematode C. elegans, chromosomes are partitioned into two chromosomal sub-regions called the “short arm” and the “long arm”: the short arm separates in the first division, and the long arm separates in the second division. The two regions themselves are determined by the position of a genetic recombination event called a crossover that normally occurs only once per chromosome. How the length of a chromosome region ultimately controls the timing of division remains a mystery. We have approached this question through a combination of quantitative microscopy imaging and physical modeling.
Project Achievements
We have identified a set of protein marks that appear on chromosomes only on the shorter of two chromosome regions. By using super-resolution fluorescence microscopy to image these marks in cases where more than two regions of different lengths occur, we have developed a model that can explain the limited appearance of these marks as the result of a feedback loop that amplifies an initial small difference in an unknown factor’s concentration between the regions. In this model, the initial difference between regions results from fast diffusion of a factor that enters the chromosome at crossovers and diffuses to both sides of the crossover, which results in the shorter region’s concentration increasing at a faster rate.
Future Prospects
While our model provides a plausible mechanism for length sensing of chromosome regions, the molecular details have yet to be confirmed. Our future work in this project will require solving problems in live imaging and further refinement of our computer simulation. The computation infrastructure and collaborations we have developed during the SPIRITS support period will continue to be of service in the elucidation of these mechanisms.
Figure
Principal Investigator

・CARLTON Peter
・Graduate School of Biostudies
・Peter Carlton is an Associate Professor in the Graduate School of Biostudies, studying chromosome dynamics in meiosis using the nematode C. elegans. He also teaches undergraduate classes in the Institute of Liberal Arts and Sciences. He has been working at Kyoto University since 2010.
